It’s hard to imagine a time when “coronavirus” wasn’t a household word. But for a long time, this family of viruses had merited very little attention. Believed to be ubiquitous among animals and avian species, the first coronavirus to infect and cause disease in humans was only isolated and identified in the 1960s.
Seven human coronaviruses have been identified since then.
Most cause only relatively minor health concerns: the common cold and seasonal respiratory infections that come around every year. But the 2003 outbreak in China and other parts of Asia of severe acute respiratory syndrome (SARS), caused by SARS-CoV (now renamed as SARS-CoV-1), propelled the virus onto the global stage. Coronaviruses gained further infamy when, in 2012, cases of the much more severe Middle East respiratory syndrome (MERS) were identified in Saudi Arabia.
Both outbreaks were relatively contained. Not surprisingly, the concern over coronavirus diseases largely faded from the minds of ordinary people. The same was true for virologists, who focused their time and funding on more pressing viruses. Then in late 2019 came SARS-CoV-2, the causative agent of COVID-19.
Fortunately, some researchers had retained an interest in coronaviruses. After all, viruses can mutate and reappear, causing new outbreaks. One such cohort, ourselves among them, works at the University of the Western Cape in South Africa. Our laboratory had, among other things, been studying some of the structural proteins that are the building blocks of coronaviruses. These proteins - named spike, nucleocapsid, membrane, and envelope proteins - have different roles, but are essential to how coronaviruses reproduce, spread and cause disease.
In our most recent paper, we examined what possibly sets the human coronaviruses that cause SARS, MERS and COVID-19 apart from the other human coronaviruses that cause milder diseases like seasonal colds. The answer, we argue, lies with the envelope protein.
Shedding light on the E protein
The envelope protein is possibly the most enigmatic and least-studied in the coronavirus-suite, owing to its small size and the difficulty of studying it in laboratory settings. In May 2019, two of us published a review paper on what was known about the envelope protein at the time.
The paper has racked up nearly 2,000 citations, most coming after the outbreak of COVID-19 – a testament less to our foresight than to the critical and previously understated role the envelope protein plays in human coronaviruses.
Even before the COVID-19 outbreak, based on what we had learnt from the SARS and MERS outbreaks, we were convinced that this protein – once written off as a “minor component” of the virus – was key to the development of disease. It is critical, for instance, in the final assembly of the virus, forming the envelope or wrapping that covers it when all its constituent components come together.
It also plays a role in the virus’s budding, when it exits from the host cell; and in the process known as pathogenesis, or the development and progression of the infection.
And it may hold a clue to either the severity or relative mildness of the disease.
Our ongoing research is beginning to suggest that the structure of the envelope protein may determine the severity of a coronavirus disease, or the difference between a blocked nose on the one hand, and collapsed lungs on the other.
The sting in the protein’s “tail”
This led us to our most recent paper. We collaborated with structural bioinformatics expert Ruben Cloete, of the South African National Bioinformatics Institute at the University of the Western Cape, to develop full-length, 3D models of the envelope proteins of five human coronaviruses: SARS-CoV-1 and -2, and MERS-CoV (responsible for the severe SARS, COVID-19 and MERS diseases); and HCoV-229E and HCoV-NL63, responsible for milder diseases. For this work, we relied on a modelling program known as MODELLER, allowing us to explore the proteins in some detail.
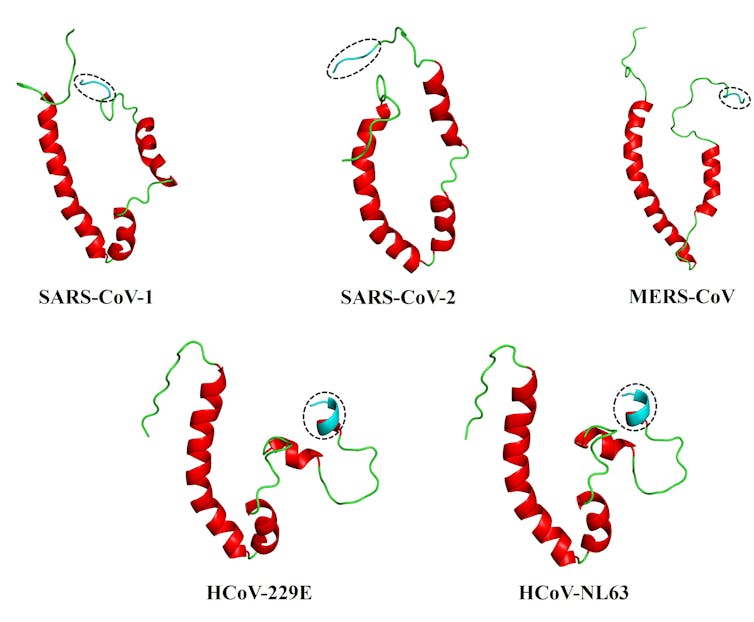
We then used a web server, HADDOCK2.4, to simulate how the envelope protein interacts with the human PALS-1 protein – an interaction already shown to be critical with SARS-CoV-1. Each of the envelope proteins could bind to the PALS-1 protein, but the coronaviruses causing SARS, MERS and COVID-19 appeared to bind more stably to PALS-1.
The answers, we believe, may lie in the conformation or shape of what’s known as the PDZ-binding motif, or PBM, which sits at the tail-end of the envelope protein. This PBM - essentially a distinctive sequence on a protein - acts like a one-of-a-kind key to a very specific lock (known as the PDZ domain) on a host cell protein. This ‘key’ allows the viral protein to interact with the host protein, making the disease worse.
We found that the more flexible, extended coil of the PBM of the coronaviruses behind SARS, MERS and COVID-19 viruses may well be what differentiates them from the more rigid PBM of the coronaviruses that cause milder diseases.
Inner workings
It is yet too early to draw definitive conclusions, as these findings will have to be confirmed with more studies – in the laboratory and in living organisms.
But it does shine some light on the inner workings of these coronaviruses and the still-enigmatic envelope protein. In so doing it could offer opportunities for the development of essential life-saving treatments and vaccines.
Dewald Schoeman received funding from National Research Foundation and the Poliomyelitis Research Foundation.
Burtram C. Fielding receives funding from the National Research Foundation.
Ruben Cloete receives funding from the Department of Higher Education and Training.
This article was originally published on The Conversation. Read the original article.







