The Nobel Prize in Chemistry was awarded on Wednesday to three scientists who have help unravel some of the enduring secrets of proteins, the building blocks of life.
While Demis Hassabis and John Jumper of Google's DeepMind lab used artificial intelligence techniques to predict the structure of proteins, biochemist David Baker managed to design totally new ones never seen in nature.
These breakthroughs are hoped to lead towards numerous advances, from discovering new drugs to enzymes that decompose pollutants.
Here is an explainer about the science behind the Nobel win.
Proteins are molecules that serve as "the factories of everything that happens in our body," Davide Calebiro, a protein researcher at the UK's University of Birmingham, told AFP.
DNA provides the blueprint for every cell. Proteins then use this information to do the work of turning that cell into something specific -- such as a brain cell or a muscle cell.
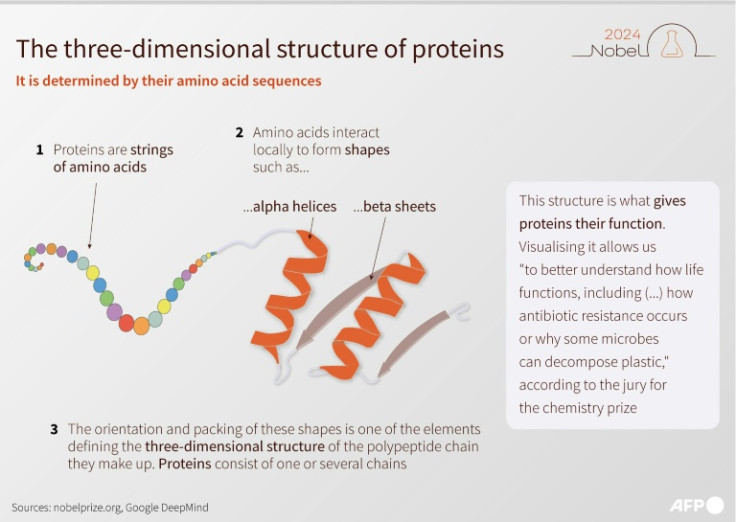
Proteins are made up of 20 different kinds of amino acid. The sequence that these acids start out in determines what 3D structure they will twist and fold into.
American Chemical Society president Mary Carroll compared how this works to an old-fashioned telephone cord.
"So you could stretch out that telephone cord, and then you would just have a one-dimensional structure," she told AFP.
"Then it would spring back" into the 3D shape, she added.
So if chemists wanted to master proteins, they needed to understand how the 2D sequences turned into these 3D structures.
"Nature already provides tens of thousands of different proteins, but sometimes we want them to do something they do not yet know how to do," said French biochemist Sophie Sacquin-Mora.
The work of previous Nobel winners had demonstrated that chemists should be able to look at amino acid sequences and predict the structure they would become.
But it was not so easy. Chemists struggled for 50 years -- there was even a biannual competition called the "Protein Olympics" where many failed the prediction test.
Enter Hassabis and Jumper. They trained their artificial intelligence model AlphaFold on all the known amino acid sequences and corresponding structures.
When given an unknown sequence, AlphaFold compares it with previous ones, gradually reconstructing the puzzle in three dimensions.
After the newer generation AlphaFold2 crushed the 2020 Protein Olympics, the organisers deemed the problem solved.
The model has now predicted the structure of almost all of the 200 million proteins known on Earth.
US biochemist Baker started at the opposite end of the process.
First, he designed an entirely new protein structure never seen in nature.
Then, using a computer programme called Rosetta that he had developed, he was able to work out the amino acid sequence that it started out as.
To achieve this, Rosetta trawled through all the known protein structures, searching for short protein fragments similar to the structure it wanted to build.
Rosetta then tweaked them and proposed a sequence that could end up as the structure.
Mastering such fundamental and important little machines as proteins could have a vast number of potential uses in the future.
"It allows us to better understand how life functions, including why some diseases develop, how antibiotic resistance occurs or why some microbes can decompose plastic," the Nobel website said.
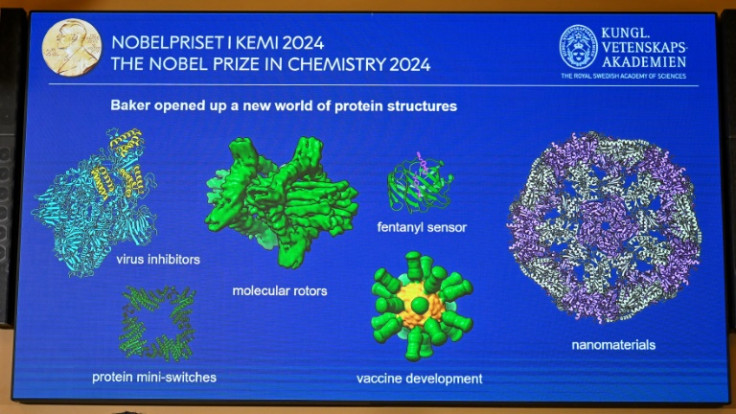
Making all-new proteins could lead to new nanomaterials, targeted drugs and vaccines, or more climate-friendly chemicals, it added.
Asked to pick a favourite protein, Baker pointed to one he "designed during the pandemic that protects against the coronavirus".
Calebiro emphasised how "transformative" this research would be.
"I think this is just the beginning of a completely new era."