
The human genome is the gift that keeps giving. We thought we had uncovered all its secrets in 2003 when scientists announced the double helix of life had been completely sequenced and assembled. To be sure, some parts were missing, but those were minor gaps dismissed as unimportant since they didn’t seem to code anything functional (what at the time was considered “junk DNA”).
But even junk has hidden treasures. Studies found variations in these unsequenced regions were intricately involved in human health, from aging to conditions like cancer and developmental disorders like autism. In 2022, a landmark study finally resolved the genomic unknown, completely sequencing the remaining eight percent of undeciphered DNA remaining.
Now, scientists are discovering that some genetic sequences encode proteins that lack any obvious ancestors, what geneticists call orphan genes. Some of these orphan genes, the researchers surmise, arose spontaneously as we evolved, unlike others that we inherited from our primate ancestors. In a paper published Tuesday in the journal Cell Reports, researchers in Ireland and Greece found around 155 of these smaller versions of DNA sequences called open reading frames (or ORF) make microproteins potentially important to a healthy cell’s growth or connected to an assortment of ailments like muscular dystrophy and retinitis pigmentosa, a rare genetic disease affecting the eyes.
“This is, I think, the first study looking at the specific evolutionary origins of these small ORFs and their microproteins,” Nikolaos Vakirlis, a scientist at the Biomedical Sciences Research Center “Alexander Fleming” in Greece and first author of the paper, tells Inverse. It’s an origin, he says, that’s been mired in much question and mystery.
Here’s the background – Most of us are familiar with our DNA consisting of four chemical building blocks called nucleotides: adenosine (A), thymine (T), cytosine (C), and guanine (G). Specific sequences, for example, AATCGA, are the recipes that ribosomes — our protein-making chefs, if you will — use to cook up a particular protein.
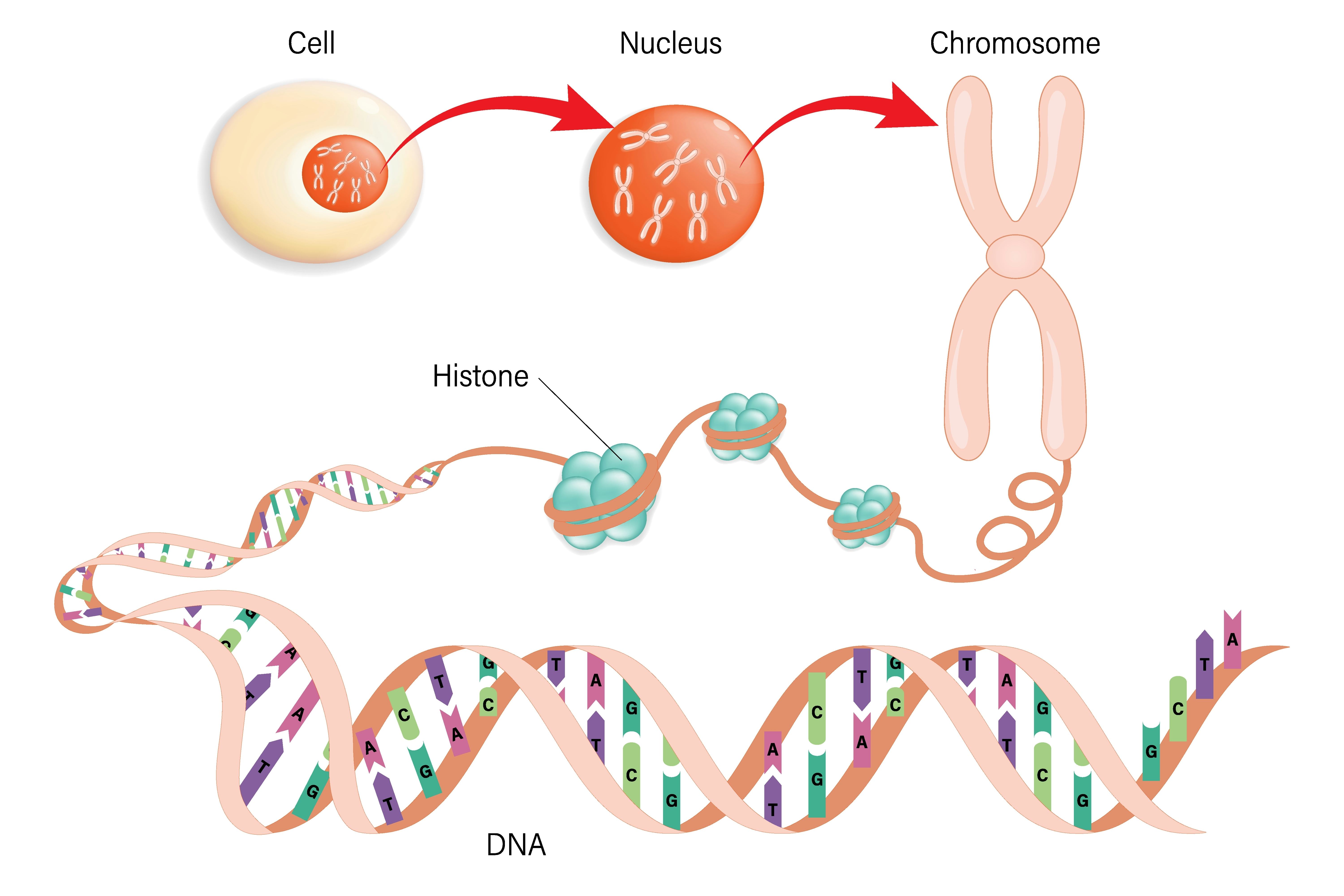
When a ribosome is reading a bunch of nucleotides, a sequence of three called a codon tells the little chef where to start and stop reading, like the first and last page of a chapter of a recipe book. The space in between the start and the stop codon is called an ORF, Aoife McLysaght, professor of genetics at Trinity College Dublin in Ireland and senior author of the paper, tells Inverse.
“It’s a piece of DNA that has the theoretical capacity to code for a protein of a certain length,” she explains. “How long it is [can vary], and we don’t know if it’s a gene without more analysis.”
The general rule of thumb scientists subscribe to is that the longer an ORF, the greater the chance it encodes a functional, meaningful protein.
“When [ORFs] are short, we basically have no way of knowing if it codes for something that’s actually biologically meaningful or if it just happens to be that way, like if you type random numbers, you’re going to come up with somebody’s phone number,” says McLysaght.
For the longest time, this led to small ORFs flying under the genomic radar, dismissed as meaningless noise. But McLysaght says there’s an emerging interest in discovering whether small ORFs are actually biologically relevant since not all genetic variation boils down to what’s conventionally considered a gene. If these small ORFs are important and make influential proteins, can they explain how new genes evolve and new traits show up in a species?
How they did it – There are countless small ORFs in the genome, so sifting through which ones are and aren’t functional and then tracing their ancestry is quite a gargantuan task.
Instead, the researchers analyzed a dataset of small ORFs in humans that were already determined to be biologically functional and pieced together a phylogenetic tree, a diagram that represents the evolutionary relationship among organisms (and quite literally looks like a tree). This was done by comparing the sequence of the small ORF against the same sequences in our close great ape relatives like the chimpanzee, orangutan, gorilla, and gibbon as well as other vertebrates. (Modern humans and apes went their separate ways about four to six million years ago, according to some estimates.)
“If you can only find the [ORF] in humans, then it appears to be human-specific,” says McLysaght. But if it’s something that’s found in humans and chimpanzees or humans, chimpanzees, and gorillas, then it could have come from a shared ancestor.
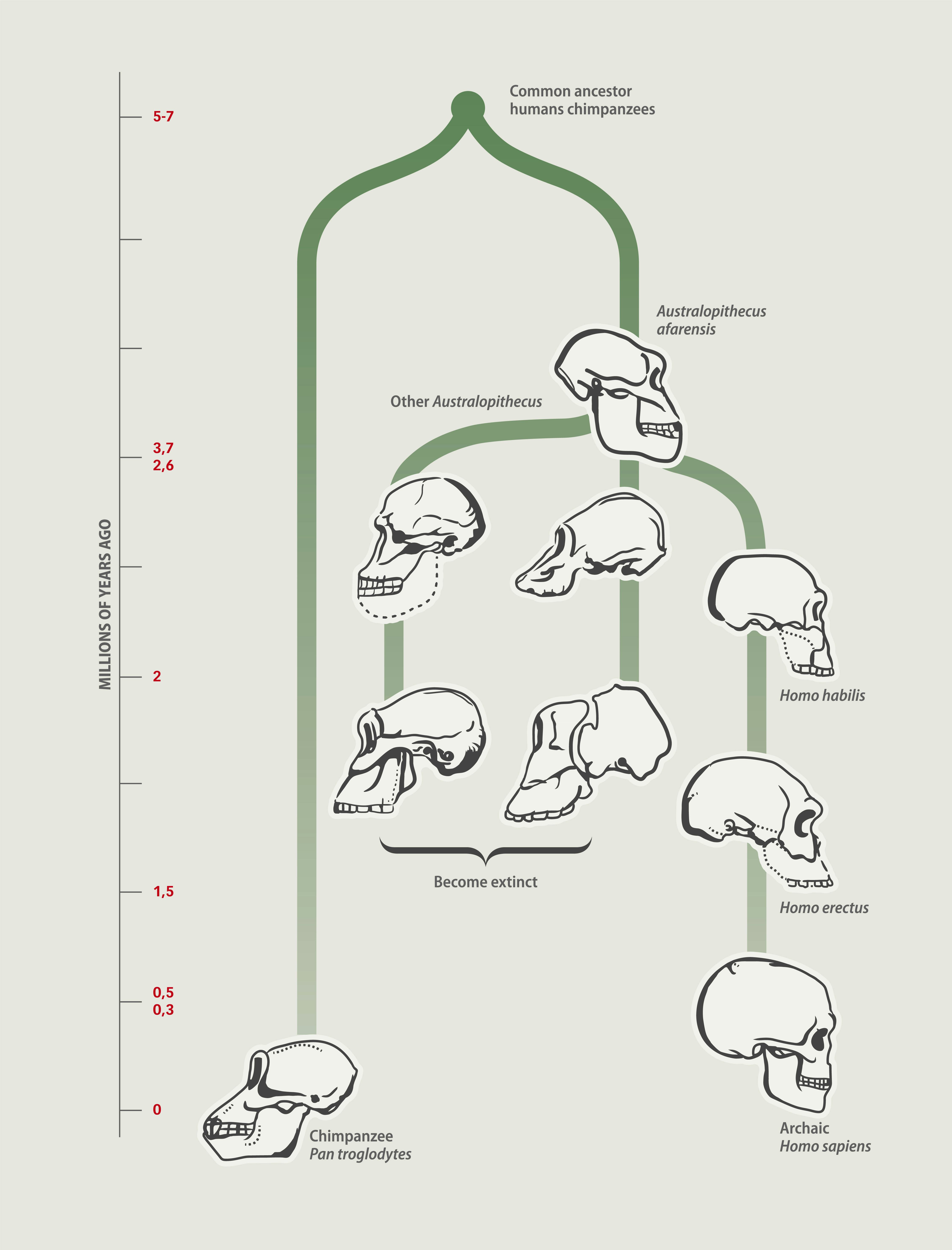
What they found — The group found that there were about 155 small ORFs producing microproteins that arose completely from scratch — what’s called de novo gene birth — in unique regions in the DNA.
A little over a quarter of these short genetic sequences — 44 exactly — appeared to be specific to humans and caused growth defects in immortalized cell lines (these are cells that ordinarily have no problem growing since they’ve got a mutation that keeps them multiplying), according to the dataset the researchers used.
Three other ORFs appear to be associated with diseases like muscular dystrophy, retinitis pigmentosa, and a rare genetic condition affecting development called Alazami syndrome that’s associated with a gene called LARP7. Another ORF seems to be strongly associated with heart tissue and is one that humans share with chimpanzees but not gorillas, orangutans, or macaques. This suggests that once an evolutionary break-up happens, genes can evolve pretty quickly within a species.
What’s next – McLysaght and Vakirilis say this is just the start to understanding how ORFs are involved in making new genes, human evolution, and their role in health and disease. (Some recent studies found these genetic sequences involved in one’s risk for Alzheimer’s disease.)
“There’s a lot more work to do and a lot more [ORFs] to study,” says Vakirlis. For instance, not every new gene confers a biological benefit. Many can be detrimental and cause harm to an organism. They don’t get passed down the generations because invariably, the organism fails to survive.
“How is that balanced with the fact we know that in the long term, entirely novel genes do emerge? How do they get over that initial, very probable situation?” says Vakirlis. “It’s a very interesting question, and I think we can address it with the data available.”







