
On March 27, 2024, a study published in the scientific journal Nature announced that the sugarcane genome had at last been cracked. This was a remarkable achievement because of the complexity of the sugarcane’s genome. That of rice, the first crop genome to be sequenced, more than 20 years ago, was “simple”: 12 chromosomes, two identical copies of each, for a total of 400 million base pairs (Mb) (nitrogen base pairs located on two complementary DNA strands).
Sugarcane is much more complex: the plant, a polyploid, contains more copies of each chromosome than conventional plants. It has 10 times more chromosomes to sequence than rice, each of them longer, for a genome that is 20 times the size. To crack the code, the CIRAD research team came up with the idea of using sorghum as a model, as it is a close cousin from the same family (Graminaceae or grasses) with far less redundancy in its genome.
But why bother studying the genome of crops and their different varieties? The diversity they contain in fact reveals the way in which they have evolved as farmers have selected them in different environments and for various purposes. For instance, rice has evolved through mutations and natural crosses between different forms that appeared around the Himalayas, which were selected each year during its domestication, which began as much as 10,000 years ago. This has resulted in a sufficient number of varieties to guarantee production in a vast range of environments.
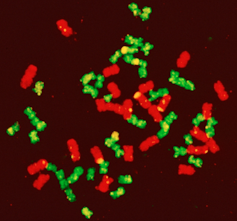
Sugarcane, for its part, comes from the islands of Southeast Asia: it is the result of broader genetic mixing, incorporating several related species. This has enabled it to combine resistance to various diseases and incomparable vegetative vigour, making it the plant that is most efficient at producing biomass. It is so tolerant of genetic mixing that all sorts of hybrids can be produced, even intergeneric ones, combining different botanical species.
Understanding how plants have adapted in the past serves to plan and speed up future crop adaptations.
Forty years of plant genome research
This is what prompted CIRAD to set up a tropical species genome analysis laboratory as long ago as 1986. The laboratory later became the Grand plateau technique régional de génotypage (regional genotyping platform). Its teams produced the first genetic maps and then, thanks to the national genotyping structure, notably the Génoscope, and to various international collaborations, they made the news in the 2010s by placing several tropical plants among the leading biological models, for instance cocoa, banana and citrus.
The list has grown longer in recent years, with arabica coffee, vanilla, and, of course, sugarcane. Progress is currently being made on coconut, rubber, oil palm, yam, groundnut and sorghum, along with fonio, extending the genome range to indigenous plants.
For plants whose varieties intercross easily, this makes it possible to pinpoint sequences associated with important agronomic characters – in the field or as regards processing and consumption. The next step is to favour them in the mixes that breeders – creators of new varieties – make before sorting progenies under growing conditions.
For other species, mixing takes longer, and is more random. It is sometimes punctuated by unlikely, almost miraculous combinations. The scientists have been able to highlight surprise hybridisations that attentive humans had managed to identify: dessert bananas, plantain bananas, oranges, lemons, grapefruit, calamondin and arabica coffee.
Cocoa, disseminated more than 5,000 years ago

To take cocoa as an example, it proved possible to study its genome diversity based on modern genomes and ancient DNA found in shards of pre-Columbian pottery.
Cocoa originated in the Amazon, and was disseminated by humans very early on, probably more than 5000 years ago, as far as the Pacific Coast and central America. Many mixes occured between populations of very distant genetic origins, which allowed cocoa to adapt to those new environments.
For chocolate fans, that mixing has also enabled the development of new aroma properties, such as those currently found in beans produced by the Criollo and Nacional varieties.
Bananas and citrus, the fruits of hybridisation
As far as bananas are concerned, genomics results suggested that their domestication began in the New Guinea region, based on hybrids between the banksii, schizocarpa and possibly zebrina groups of the species Musa acuminata.
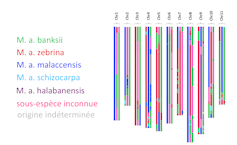
Those first cultivars were then transported to different regions of Southeast Asia and hybridised with other local sub-species of the genus Musa, resulting in their diversification and in the different types of varieties we now know, which involve up to seven ancestral contributors.
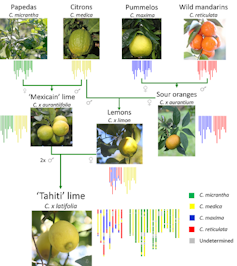
As for citrus species, their genome diversity shows that most of the species grown in modern times result from four founding species. For instance, limes (Citrus latifolia) are the result of natural hybridisation between the Mediterranean lemon and the key lime, and involve four ancestral species.
Genomics has shed light on the way in which reproductive cells (gametes) are produced in these complex genomic contexts, and can help to pave the way for breeding disease-resistant rootstocks.
Adapting coffee
Lastly, arabica coffee is also the result of one of these unlikely cases of hybridisation. It fused the genomes of two different species (Coffea canephora and C. eugenioides), but this only ever happened once.
This new cross occurred some 500,000 years ago in Ethiopia and the Arabs chose it to produce coffee as of the 14th century or thereabouts. It brought many properties as a result of the diversity within each plant, but all the plants derived from it are almost identical.
From then on, to continue its adaptation, seeking out other rare events that could broaden that diversity through introgression from other sources, became the priority. Attention is now focusing on localised forms in Ethiopia and Yemen.
Precious resources for genetic improvement
All these unlikely events specific to each crop were very beneficial, but are difficult to reproduce. It will not be possible to explore variants in order to create and maintain the diversity required for adaptation until we understand all the details. Certain forms, some of them extremely rare, are the only sources of characters that could be crucially important in future.
CIRAD, in collaboration with INRAE and IRD and in association with international research networks and private structures, conserves and maintains genetic resource collections, in the form of seeds, freeze-dried cells or whole plants, in Montpellier, Corsica, Guadeloupe, French Guiana and Réunion.
These activities are costly, but represent an investment that is essential for the future. By mobilising the available diversity, in close association with other players – from farmers to research centres – we are preparing the varieties of tomorrow.
Genomic diversity, a key factor in future action, has become a global issue. If we know how to reveal what certain sequences have to offer in terms of biological potential, we can steer their transmission to progenies. It may even be possible to rewrite them by means of genome editing, to transmit desirable biological attributes to existing varieties without having to resort to crossing.
Provided the biotechnical knowledge exists, this genetic improvement method appears limitless, notably by facilitating mixing. As a result, issues surrounding societal choices and intellectual property are now emerging in a knowledge field that is currently a subject for great debate.
Disseminating knowledge
First and foremost, this exploration of genome diversity is in response to an environmental challenge, climate change, which necessitates speeding up operations to adapt plants to specific contexts – different growing zones –, both contrasting and fast-changing, in close interaction with producers.
However, it also addresses a societal issue: it is essential to guarantee access to information, to enable everyone to take it on board, adapt their crops and continue to feed themselves in a sustainable way. We are therefore working to broaden access to data and analysis tools, by means of bioinformatics platforms housing dedicated genome databases for each plant. And we are helping to build fairer skillsets by providing training in functional and comparative genomics tools and how to use them, for junior scientists in France and at our partners in the global South.
The authors would like to thank their colleagues, in particular Angélique D'hont, Patrick Ollitrault and Benoît Bertrand, as well as Francis Quétier from Génoscope, for their pioneering and decisive contributions and their founding role.
CIRAD was founded by decree on 5 June 1984. For the past 40 years, CIRAD scientists have been sharing and co-constructing knowledge and innovative solutions with developing countries to preserve biodiversity and plant and animal health, and thus make agricultural and food systems more sustainable and resilient in the face of global change.
Jean Christophe Glaszmann has received funding from the ANR, the European Union and the CGIAR.
Claire Billot has received funding from the ANR (France), the European Union, the CGIAR and CORAF (Africa).
Claire Lanaud has received funding from the French National Research Agency (ANR), the Agropolis Foundation, MUSE (Montpellier University of Excellence) and private chocolate manufacturers such as Valrhona, which are interested in the diversity of cocoa tree genetic resources.
This article was originally published on The Conversation. Read the original article.







